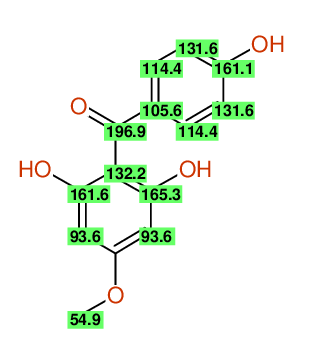
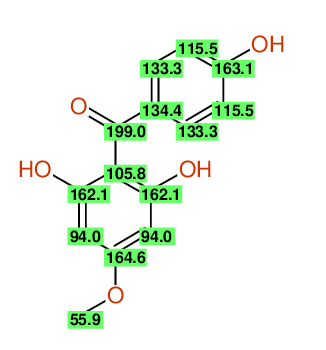
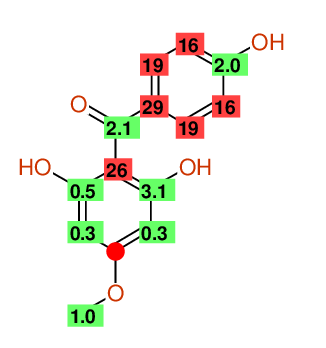
Compound: 2_,6_,4-TRIHYDROXY-4-METHOXYBENZOPHENONE
Project: MOLECULES,20,4410[2015]CSEARCH-NUMBER:UWPA003032-20210427
Did you know ? |
Your query structure is automatically matched against the PUBCHEM-collection, |
|
Request from: molecules@robien.at Compound: 2_,6_,4-TRIHYDROXY-4-METHOXYBENZOPHENONE Project: MOLECULES,20,4410[2015]CSEARCH-NUMBER:UWPA003032-20210427 |
 |
|
Compound: 2_,6_,4-TRIHYDROXY-4-METHOXYBENZOPHENONE Project: MOLECULES,20,4410[2015]CSEARCH-NUMBER:UWPA003032-20210427 | 
|

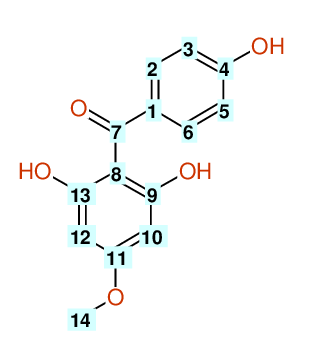
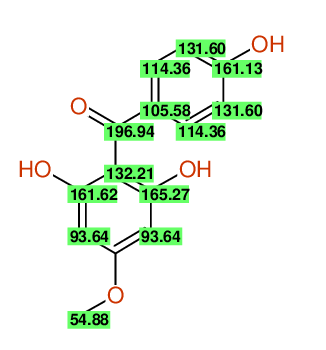
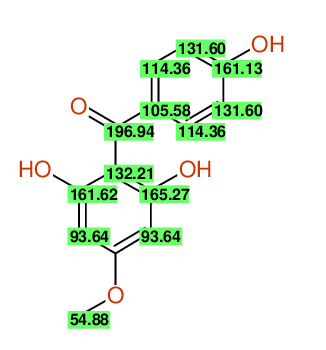
| Carbon number | Chemical Shift Value | Multiplicity from Structure | Multiplicity from Experiment |
| 1 | 105.58 | S | - |
| 2 | 114.36 | D | - |
| 3 | 131.60 | D | - |
| 4 | 161.13 | S | - |
| 5 | 131.60 | D | - |
| 6 | 114.36 | D | - |
| 7 | 196.94 | S | - |
| 8 | 132.21 | S | - |
| 9 | 165.27 | S | - |
| 10 | 93.64 | D | - |
| 12 | 93.64 | D | - |
| 13 | 161.62 | S | - |
| 14 | 54.88 | Q | - |
| Checking lines & multiplicity | Carbons/Lines | Singlet | Dublet | Triplet | Quartet | Odd | Even | None |
| From structure | 14 | 7 | 6 | 0 | 1 | 7 | 7 | 0 |
| From spectrum | 13 | 6 | 6 | 0 | 1 | 6 | 7 | 0 |

|
|
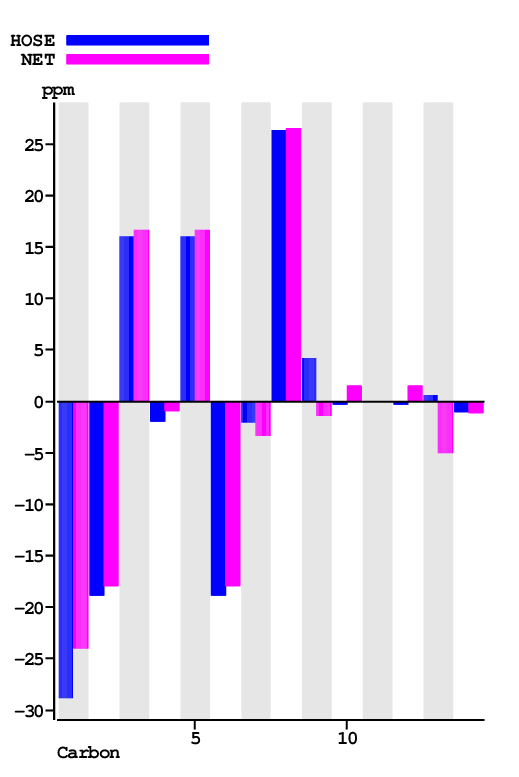
| Carbon Number | Neural Network Prediction |
HOSE-Code Prediction |
Preferred Value from both Predictions |
Experimental values |
Difference (Exp-Pred/ppm) |
Assignment | Prediction Quality |
|---|---|---|---|---|---|---|---|
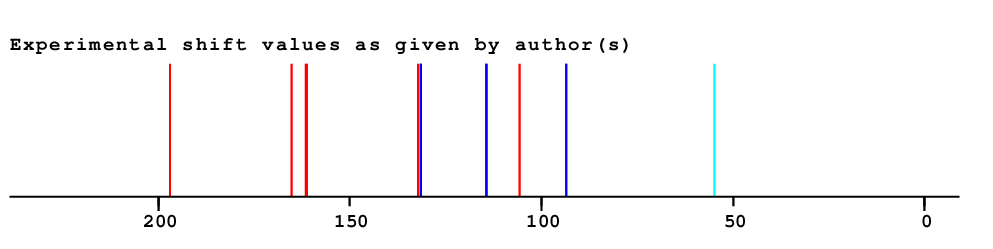
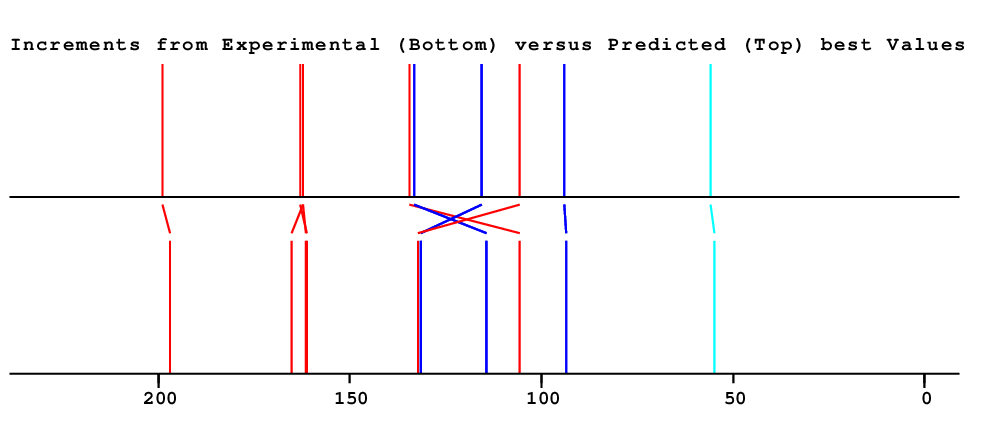
| 1 | 129.6 | 134.4 | 134.4 | 105.6 | 28.8 | Assigned by author | Only very few similar structures |
| 2 | 132.3 | 133.3 | 133.3 | 114.4 | 18.9 | Assigned by author | |
| 3 | 115.0 | 115.5 | 115.5 | 131.6 | 16.1 | Assigned by author | |
| 4 | 162.1 | 163.1 | 163.1 | 161.1 | 2.0 | Assigned by author | |
| 5 | 115.0 | 115.5 | 115.5 | 131.6 | 16.1 | Assigned by author | |
| 6 | 132.3 | 133.3 | 133.3 | 114.4 | 18.9 | Assigned by author | |
| 7 | 200.3 | 199.0 | 199.0 | 196.9 | 2.1 | Assigned by author | |
| 8 | 105.6 | 105.8 | 105.8 | 132.2 | 26.4 | Assigned by author | |
| 9 | 166.7 | 161.0 | 162.1 | 165.3 | 3.1 | Assigned by author |
Large Difference between NET & HOSE Only very few similar structures |
| 10 | 92.1 | 94.0 | 94.0 | 93.6 | 0.3 | Assigned by author | |
| 11 | 166.9 | 164.6 | 164.6 | Chemical shift not available | |||
| 12 | 92.1 | 94.0 | 94.0 | 93.6 | 0.3 | Assigned by author | |
| 13 | 166.7 | 161.0 | 162.1 | 161.6 | 0.5 | Assigned by author |
Large Difference between NET & HOSE Only very few similar structures |
| 14 | 56.0 | 55.9 | 55.9 | 54.9 | 1.0 | Assigned by author |

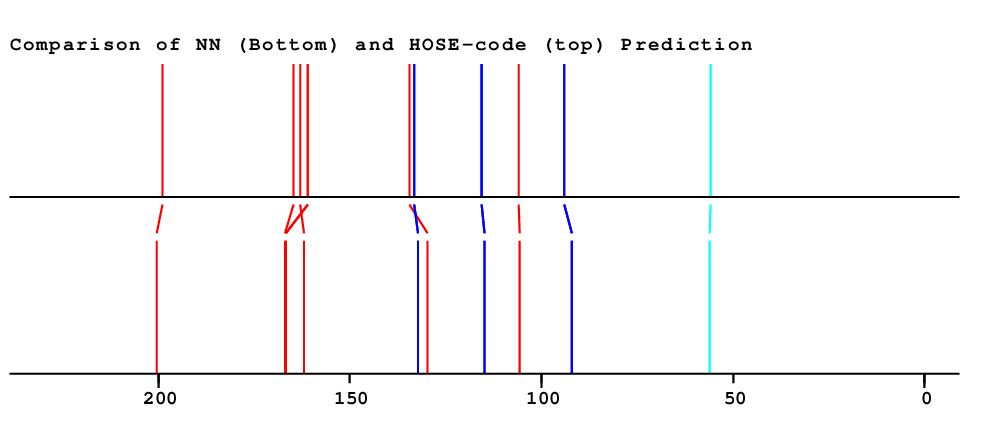
| HOSE | NET | NET&HOSE | NONE |




| ||
 |
 |

|

Compound: 2_,6_,4-TRIHYDROXY-4-METHOXYBENZOPHENONE |

|
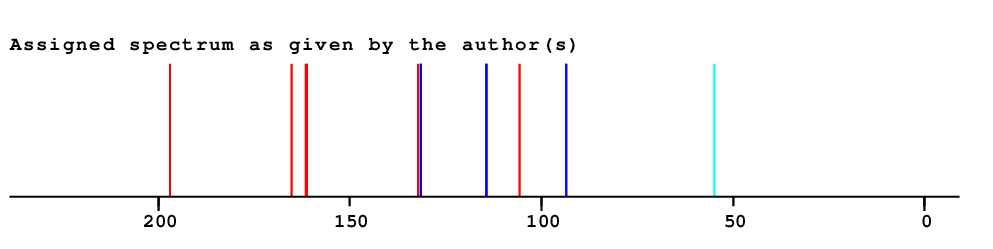
| Experimental values | ||||
 | ||||
| Symmetry considerations | ||||
 | ||||
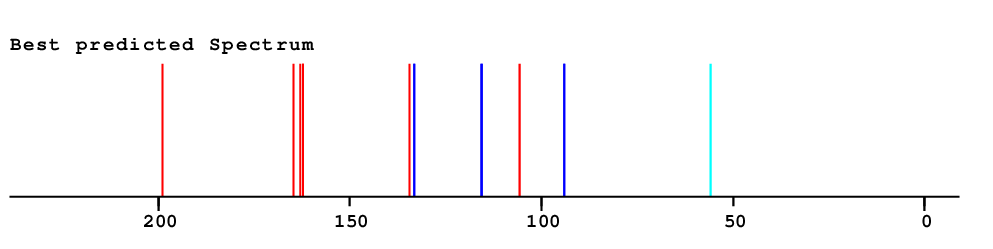
| Predicted values | ||||
 | ||||
| Matching map | ||||
 | ||||
| Deviation per position ( Average is 10.3ppm ) | ||||
 | ||||
| ||||
 | ||||
| Overall Similarity Index is 13.6 0.0 is a "perfect match", up to approximately 3.0 it is "reasonable", above 5.0 it is more or less "unbelievable" |

| ||
 |
 |
686 Requests have been launched by molecules@robien.at | |||||
Year |
Accept |
Minor Revision |
Major Revision |
Reject | Only Prediction |
| 2021 | 36 | 453 | 166 | 31 | |