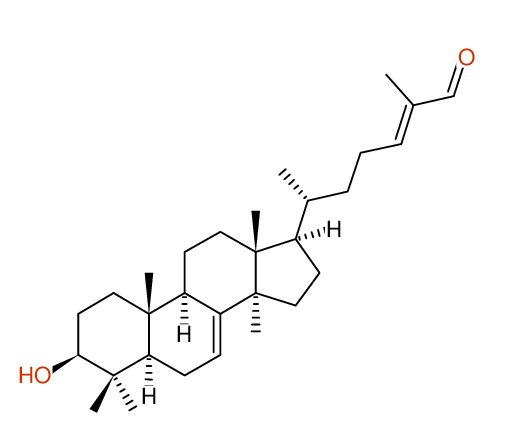
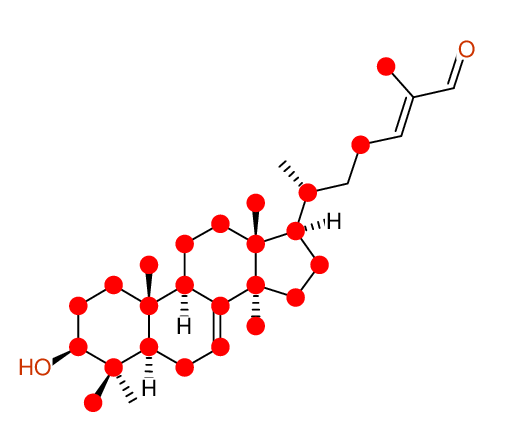
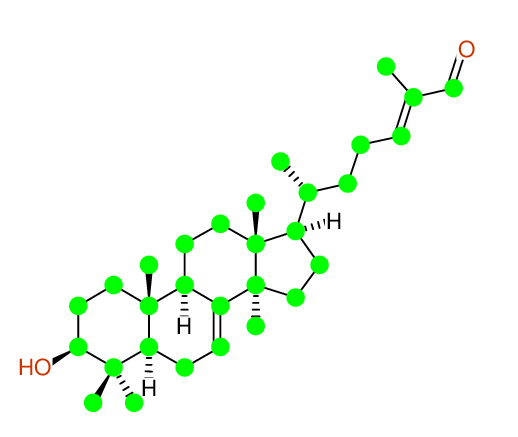
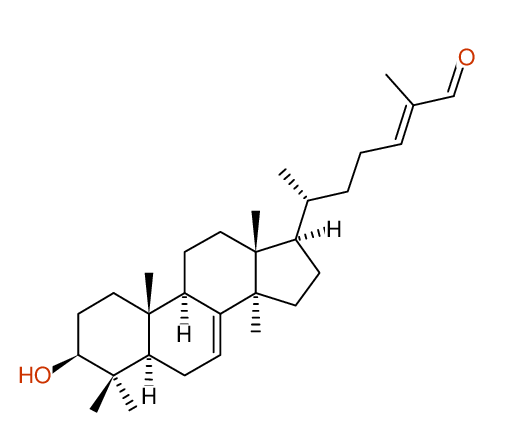
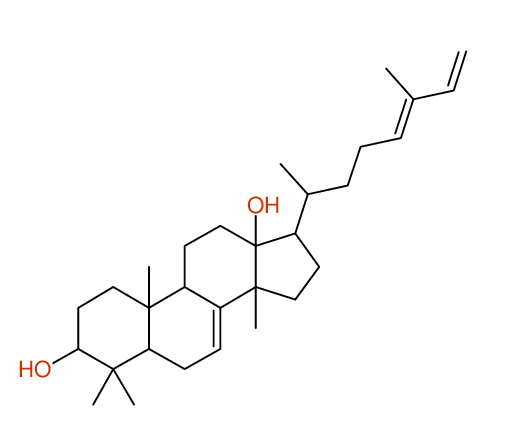
Compound: KIUSIANIN-A_3-BETA-HYDROXY-LANOSTANE-7,[24E]-DIEN-26-AL_COMPOUND-1
Project: CHEM.PHARM.BULL.,62,937[2014]CSEARCH-NUMBER:UWBT013934-20160521
Did you know ? |
You can build your own database from previous requests. |
|
Request from: cpb.kiusianin@robien.org Compound: KIUSIANIN-A_3-BETA-HYDROXY-LANOSTANE-7,[24E]-DIEN-26-AL_COMPOUND-1 Project: CHEM.PHARM.BULL.,62,937[2014]CSEARCH-NUMBER:UWBT013934-20160521 |
 |
|
Compound: KIUSIANIN-A_3-BETA-HYDROXY-LANOSTANE-7,[24E]-DIEN-26-AL_COMPOUND-1 Project: CHEM.PHARM.BULL.,62,937[2014]CSEARCH-NUMBER:UWBT013934-20160521 | 
|

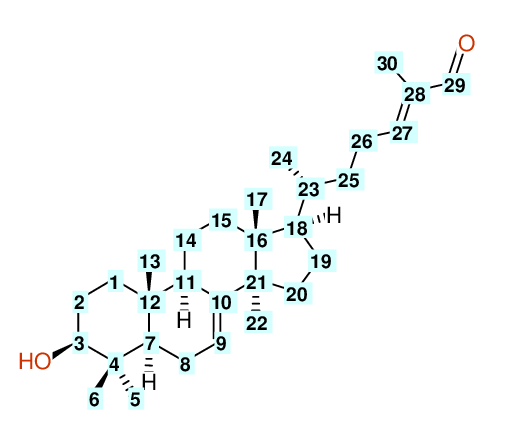
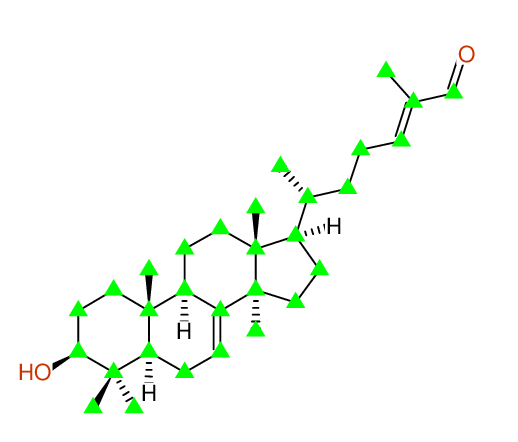
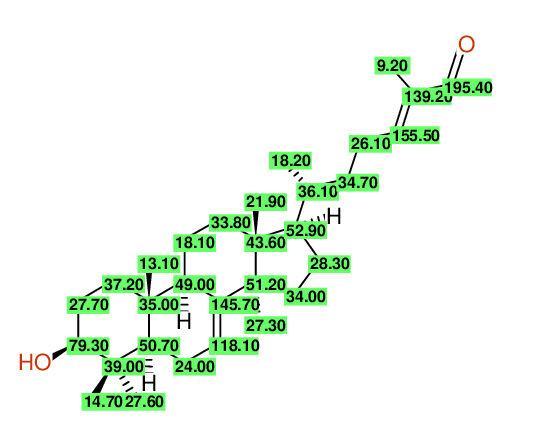
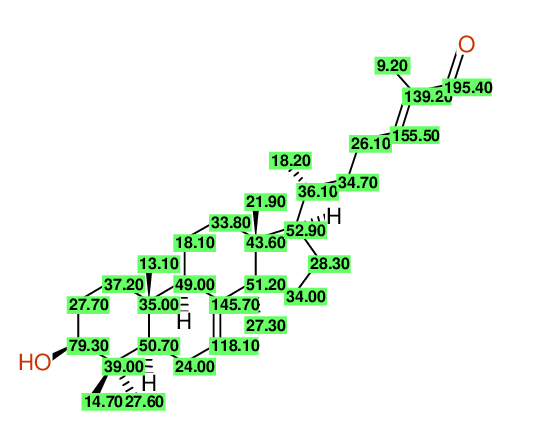
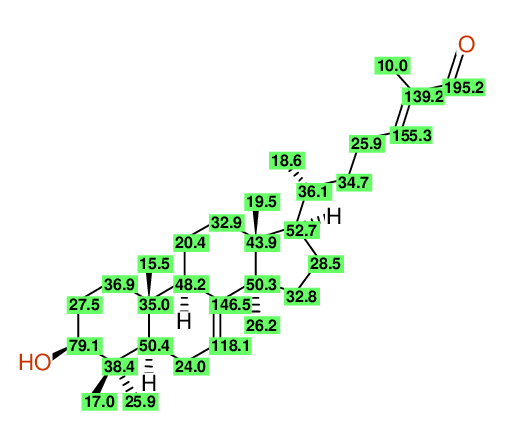
| Carbon number | Chemical Shift Value | Multiplicity from Structure | Multiplicity from Experiment |
| 1 | 37.20 | T | - |
| 2 | 27.70 | T | - |
| 3 | 79.30 | D | - |
| 4 | 39.00 | S | - |
| 5 | 27.60 | Q | - |
| 6 | 14.70 | Q | - |
| 7 | 50.70 | D | - |
| 8 | 24.00 | T | - |
| 9 | 118.10 | D | - |
| 10 | 145.70 | S | - |
| 11 | 49.00 | D | - |
| 12 | 35.00 | S | - |
| 13 | 13.10 | Q | - |
| 14 | 18.10 | T | - |
| 15 | 33.80 | T | - |
| 16 | 43.60 | S | - |
| 17 | 21.90 | Q | - |
| 18 | 52.90 | D | - |
| 19 | 28.30 | T | - |
| 20 | 34.00 | T | - |
| 21 | 51.20 | S | - |
| 22 | 27.30 | Q | - |
| 23 | 36.10 | D | - |
| 24 | 18.20 | Q | - |
| 25 | 34.70 | T | - |
| 26 | 26.10 | T | - |
| 27 | 155.50 | D | - |
| 28 | 139.20 | S | - |
| 29 | 195.40 | D | - |
| 30 | 9.20 | Q | - |
| Checking lines & multiplicity | Carbons/Lines | Singlet | Dublet | Triplet | Quartet | Odd | Even | None |
| From structure | 30 | 6 | 8 | 9 | 7 | 15 | 15 | 0 |
| From spectrum | 30 | 6 | 8 | 9 | 7 | 15 | 15 | 0 |

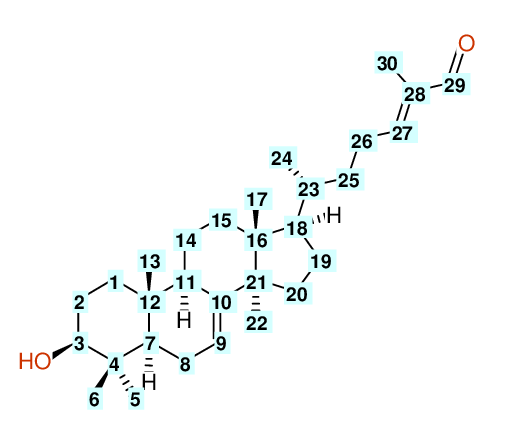
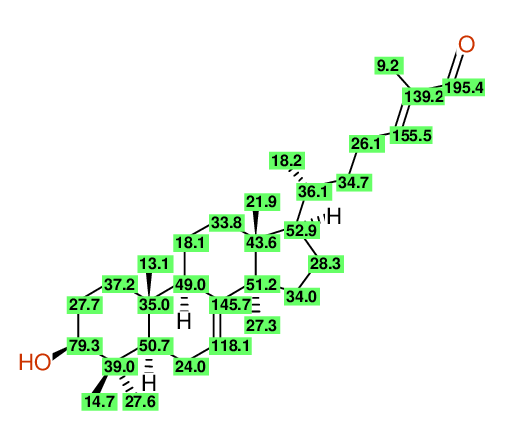
| Carbon Number | Neural Network Prediction |
HOSE-Code Prediction |
Preferred Value from both Predictions |
Experimental values |
Difference (Exp-Pred/ppm) |
Assignment | Prediction Quality |
|---|---|---|---|---|---|---|---|
| 1 | 37.3 | 36.9 | 36.9 | 37.2 | 0.3 | Assigned by author | |
| 2 | 27.4 | 27.5 | 27.5 | 27.7 | 0.2 | Assigned by author Check assignment - maybe 27.60 ? |
|
| 3 | 78.8 | 79.1 | 79.1 | 79.3 | 0.2 | Assigned by author | |
| 4 | 38.1 | 38.4 | 38.4 | 39.0 | 0.6 | Assigned by author | |
| 5 | 25.5 | 25.9 | 25.9 | 27.6 | 1.7 | Assigned by author Check assignment - maybe 27.30 ? |
|
| 6 | 16.8 | 17.0 | 17.0 | 14.7 | 2.3 | Assigned by author Check assignment - maybe 18.10 ? |
Inconsistent assignments in reference material |
| 7 | 50.3 | 50.4 | 50.4 | 50.7 | 0.3 | Assigned by author | |
| 8 | 24.1 | 24.0 | 24.0 | 24.0 | 0.0 | Assigned by author | |
| 9 | 120.7 | 118.1 | 118.1 | 118.1 | 0.0 | Assigned by author | Inconsistent assignments in reference material |
| 10 | 144.4 | 146.5 | 146.5 | 145.7 | 0.8 | Assigned by author | |
| 11 | 45.4 | 48.2 | 48.2 | 49.0 | 0.8 | Assigned by author | |
| 12 | 36.2 | 35.0 | 35.0 | 35.0 | 0.0 | Assigned by author | |
| 13 | 16.5 | 15.5 | 15.5 | 13.1 | 2.4 | Assigned by author Check assignment - maybe 14.70 ? |
Inconsistent assignments in reference material |
| 14 | 22.0 | 20.4 | 20.4 | 18.1 | 2.3 | Assigned by author Check assignment - maybe 21.90 ? |
Inconsistent assignments in reference material |
| 15 | 31.3 | 32.9 | 32.9 | 33.8 | 0.9 | Assigned by author | |
| 16 | 44.9 | 43.9 | 43.9 | 43.6 | 0.3 | Assigned by author | |
| 17 | 16.4 | 19.5 | 19.5 | 21.9 | 2.4 | Assigned by author Check assignment - maybe 13.10 ? |
Inconsistent assignments in reference material |
| 18 | 50.4 | 52.7 | 52.7 | 52.9 | 0.2 | Assigned by author | |
| 19 | 28.5 | 28.5 | 28.5 | 28.3 | 0.2 | Assigned by author | |
| 20 | 31.3 | 32.8 | 32.8 | 34.0 | 1.2 | Assigned by author | |
| 21 | 51.9 | 50.3 | 50.3 | 51.2 | 0.9 | Assigned by author | |
| 22 | 24.8 | 26.2 | 26.2 | 27.3 | 1.1 | Assigned by author Check assignment - maybe 26.10 ? |
|
| 23 | 36.4 | 36.1 | 36.1 | 36.1 | 0.0 | Assigned by author | |
| 24 | 18.7 | 18.6 | 18.6 | 18.2 | 0.4 | Assigned by author | |
| 25 | 35.0 | 34.7 | 34.7 | 34.7 | 0.0 | Assigned by author | |
| 26 | 26.4 | 25.9 | 25.9 | 26.1 | 0.2 | Assigned by author Check assignment - maybe 27.70 ? |
|
| 27 | 156.7 | 155.3 | 155.3 | 155.5 | 0.2 | Assigned by author | |
| 28 | 137.4 | 139.2 | 139.2 | 139.2 | 0.0 | Assigned by author | |
| 29 | 192.9 | 195.2 | 195.2 | 195.4 | 0.2 | Assigned by author | Inconsistent assignments in reference material |
| 30 | 9.5 | 10.0 | 10.0 | 9.2 | 0.8 | Assigned by author |
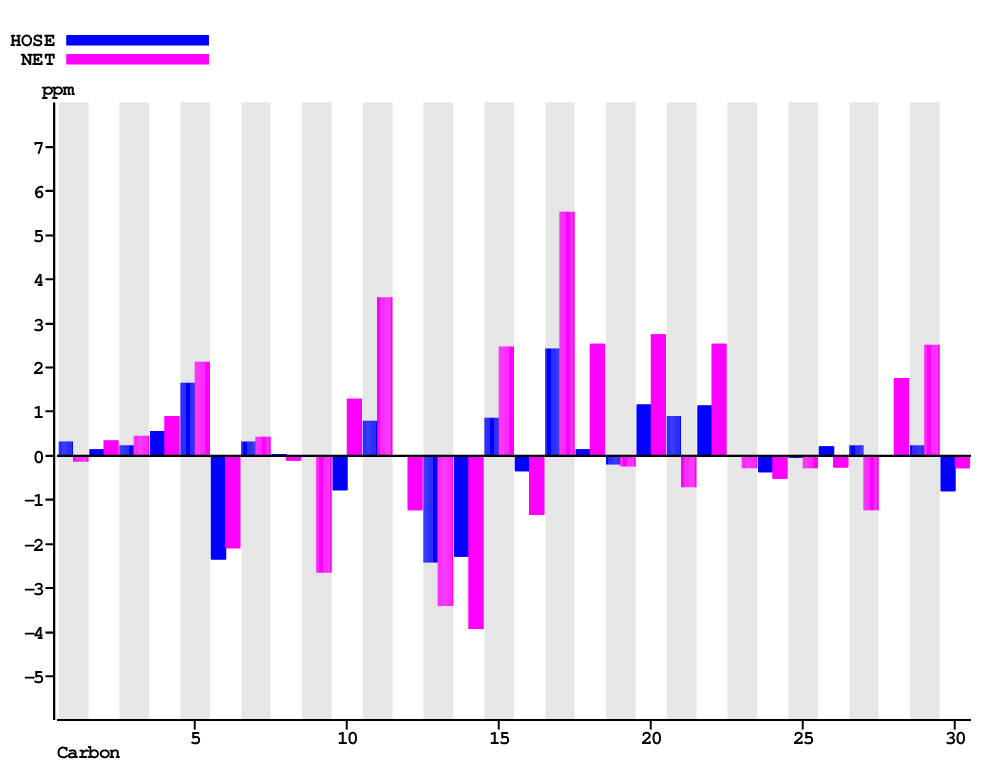

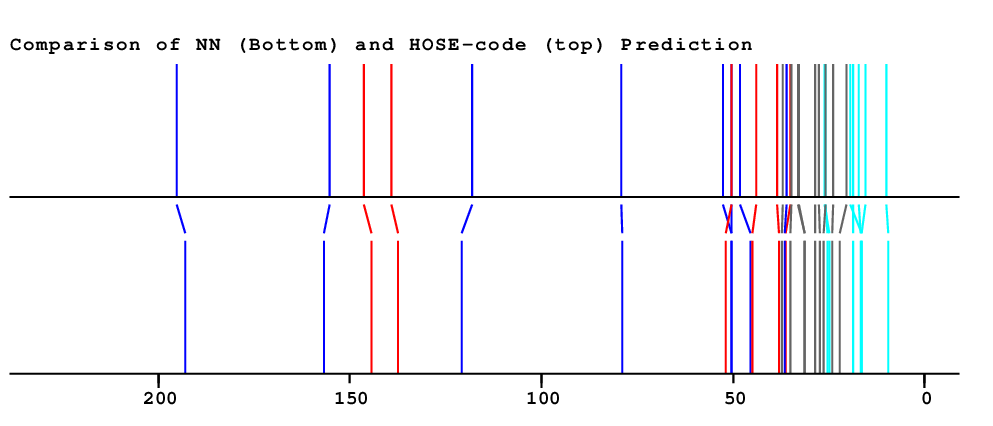
| HOSE | NET | NET&HOSE | NONE |



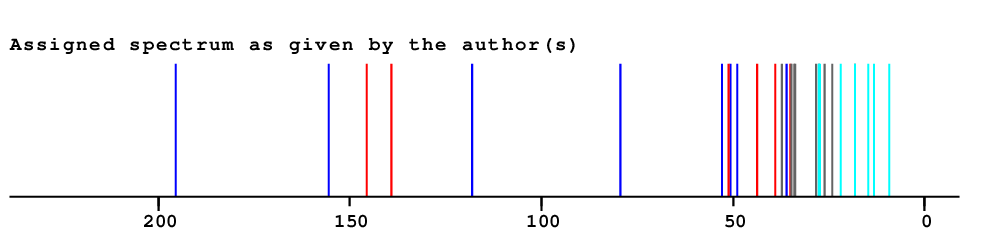

| ||
 |
 |
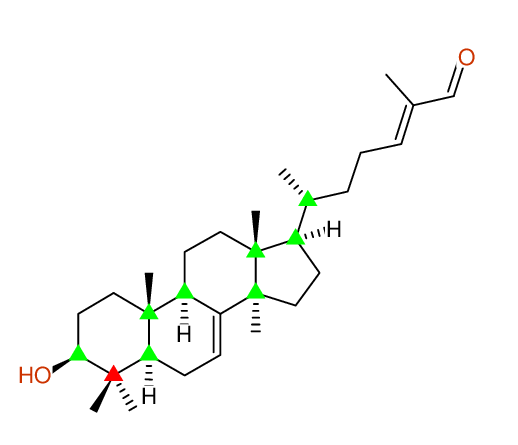
| Identical structure: Entry # 1 of 1 |
CHEM.PHARM.BULL.,62,937(2014) |
|
| Solvent: CDCL3 30 carbons given 30 lines given 30 assigned lines 0 exchangeable shifts C30H48O2 |
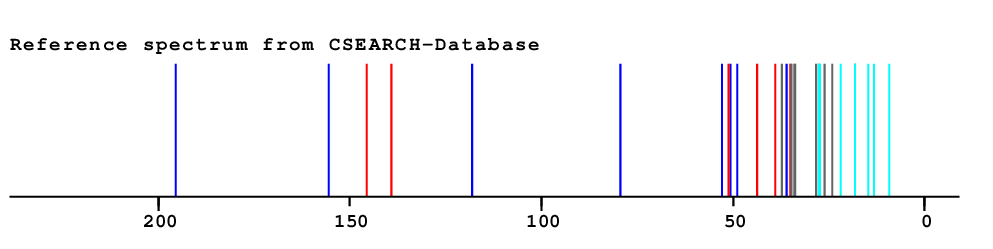 |
|
CXPKYCXFLMPMEZ Search Structure on WEB Availability of spectral data  |
 |
|

|

Your evaluation was classified either as
"Major revision" or "Reject", |

|
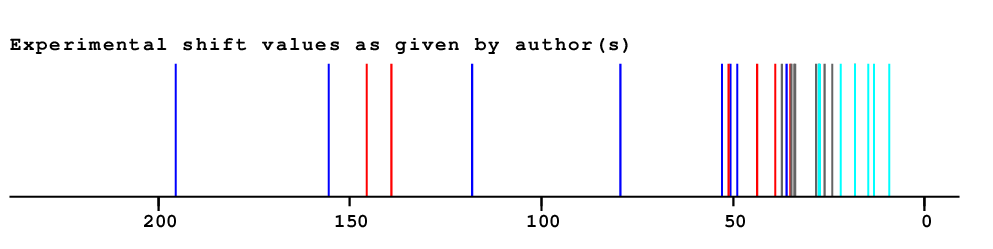
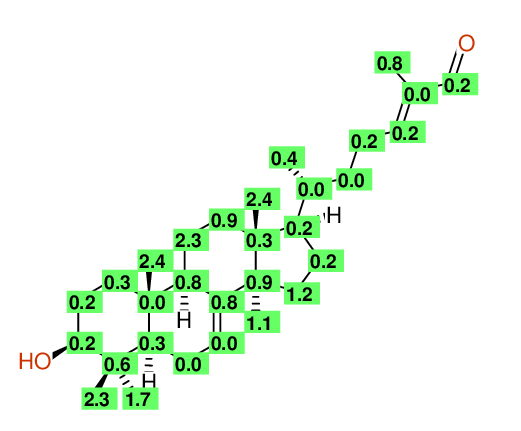
| Experimental values | ||||
 | ||||
| Predicted values | ||||
 | ||||
| Matching map | ||||
 | ||||
| Deviation per position ( Average is 0.7ppm ) | ||||
 | ||||
 | ||||
| ||||
 | ||||
| Overall Similarity Index is 1.2 0.0 is a "perfect match", up to approximately 3.0 it is "reasonable", above 5.0 it is more or less "unbelievable" |

| ||
 |
 |

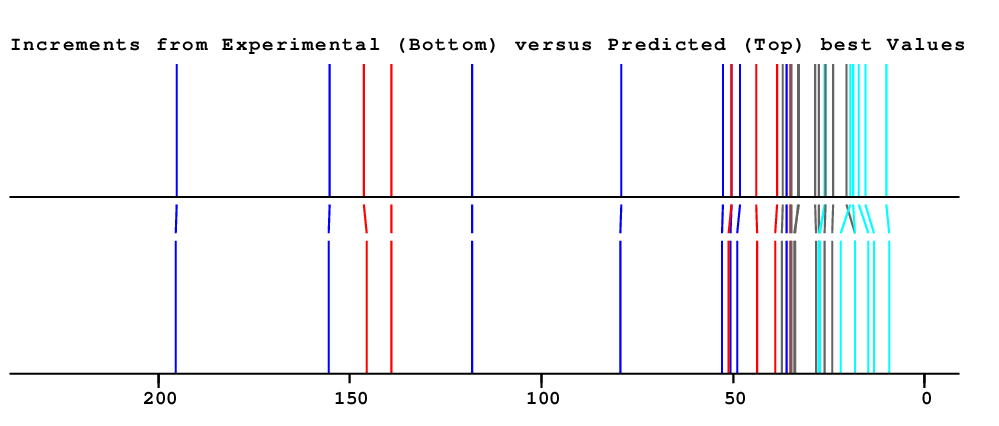
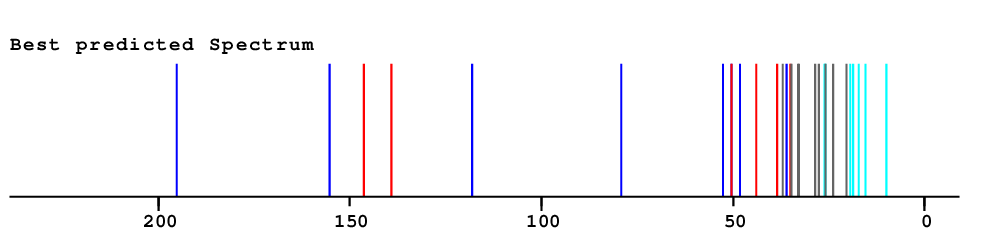
| Carbon | Experimental shift value |
NN-Value | Difference to experimental value |
HOSE-Value | Difference to experimental value |
Best-Value | Difference to experimental value |
Total Deviation |
| 1 | 37.20 | 37.33 | 0.13 | 36.89 | 0.31 | 36.89 | 0.31 | 0.76 |
| 2 | 27.70 | 27.36 | 0.34 | 27.55 | 0.15 | 27.55 | 0.15 | 0.64 |
| 3 | 79.30 | 78.84 | 0.46 | 79.06 | 0.24 | 79.06 | 0.24 | 0.93 |
| 4 | 39.00 | 38.10 | 0.90 | 38.43 | 0.57 | 38.43 | 0.57 | 2.03 |
| 5 | 27.60 | 25.47 | 2.13 | 25.95 | 1.65 | 25.95 | 1.65 | 5.44 |
| 6 | 14.70 | 16.79 | 2.09 | 17.04 | 2.34 | 17.04 | 2.34 | 6.77 |
| 7 | 50.70 | 50.28 | 0.42 | 50.39 | 0.31 | 50.39 | 0.31 | 1.05 |
| 8 | 24.00 | 24.12 | 0.12 | 23.96 | 0.04 | 23.96 | 0.04 | 0.20 |
| 9 | 118.10 | 120.75 | 2.65 | 118.12 | 0.02 | 118.12 | 0.02 | 2.70 |
| 10 | 145.70 | 144.41 | 1.29 | 146.49 | 0.79 | 146.49 | 0.79 | 2.86 |
| 11 | 49.00 | 45.41 | 3.59 | 48.20 | 0.80 | 48.20 | 0.80 | 5.18 |
| 12 | 35.00 | 36.22 | 1.22 | 35.00 | 0.00 | 35.00 | 0.00 | 1.22 |
| 13 | 13.10 | 16.51 | 3.41 | 15.51 | 2.41 | 15.51 | 2.41 | 8.22 |
| 14 | 18.10 | 22.02 | 3.92 | 20.39 | 2.29 | 20.39 | 2.29 | 8.50 |
| 15 | 33.80 | 31.32 | 2.48 | 32.95 | 0.85 | 32.95 | 0.85 | 4.18 |
| 16 | 43.60 | 44.93 | 1.33 | 43.94 | 0.34 | 43.94 | 0.34 | 2.00 |
| 17 | 21.90 | 16.38 | 5.52 | 19.47 | 2.43 | 19.47 | 2.43 | 10.39 |
| 18 | 52.90 | 50.36 | 2.54 | 52.75 | 0.15 | 52.75 | 0.15 | 2.84 |
| 19 | 28.30 | 28.53 | 0.23 | 28.50 | 0.20 | 28.50 | 0.20 | 0.64 |
| 20 | 34.00 | 31.25 | 2.75 | 32.84 | 1.16 | 32.84 | 1.16 | 5.07 |
| 21 | 51.20 | 51.92 | 0.72 | 50.30 | 0.90 | 50.30 | 0.90 | 2.52 |
| 22 | 27.30 | 24.77 | 2.53 | 26.17 | 1.13 | 26.17 | 1.13 | 4.80 |
| 23 | 36.10 | 36.39 | 0.29 | 36.10 | 0.00 | 36.10 | 0.00 | 0.29 |
| 24 | 18.20 | 18.73 | 0.53 | 18.57 | 0.37 | 18.57 | 0.37 | 1.26 |
| 25 | 34.70 | 34.98 | 0.28 | 34.73 | 0.03 | 34.73 | 0.03 | 0.35 |
| 26 | 26.10 | 26.36 | 0.26 | 25.88 | 0.22 | 25.88 | 0.22 | 0.70 |
| 27 | 155.50 | 156.72 | 1.22 | 155.27 | 0.23 | 155.27 | 0.23 | 1.67 |
| 28 | 139.20 | 137.43 | 1.77 | 139.19 | 0.01 | 139.19 | 0.01 | 1.79 |
| 29 | 195.40 | 192.88 | 2.52 | 195.17 | 0.23 | 195.17 | 0.23 | 2.98 |
| 30 | 9.20 | 9.48 | 0.28 | 9.99 | 0.79 | 9.99 | 0.79 | 1.87 |
| 1.60 | 0.70 | 0.70 | 3.00 |
|
| |
|
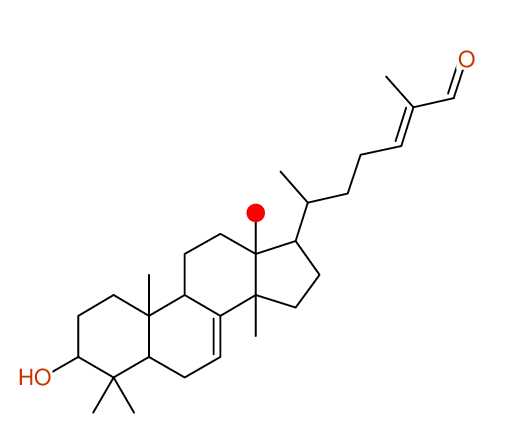
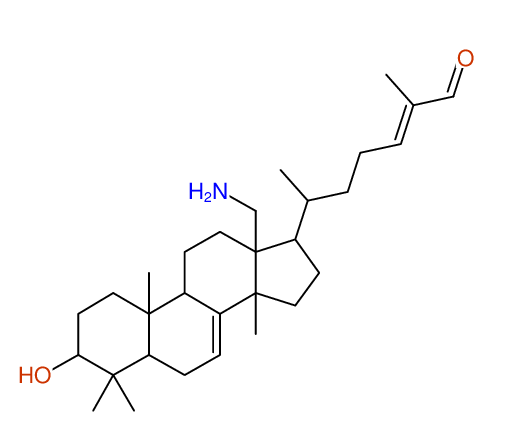
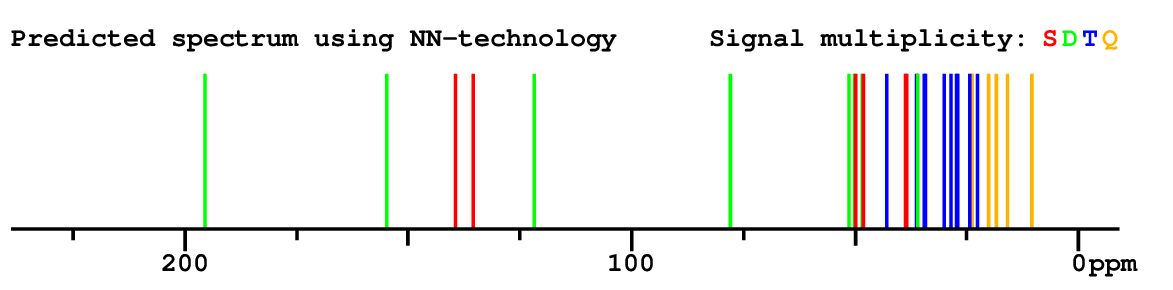
Structure Proposal # 1 ZPHBLIHVNGZGER Deviation = 1.35ppm
Search WEB for skeleton respect to your proposal |
|
|
|
|
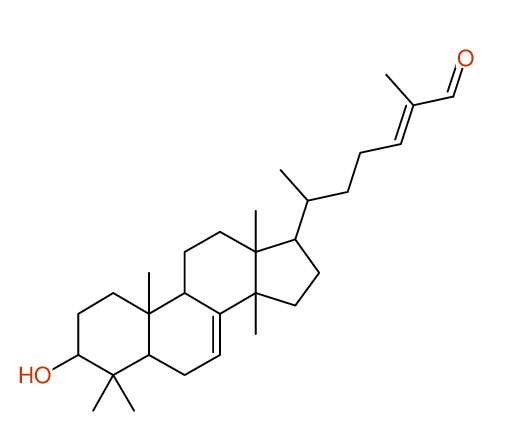
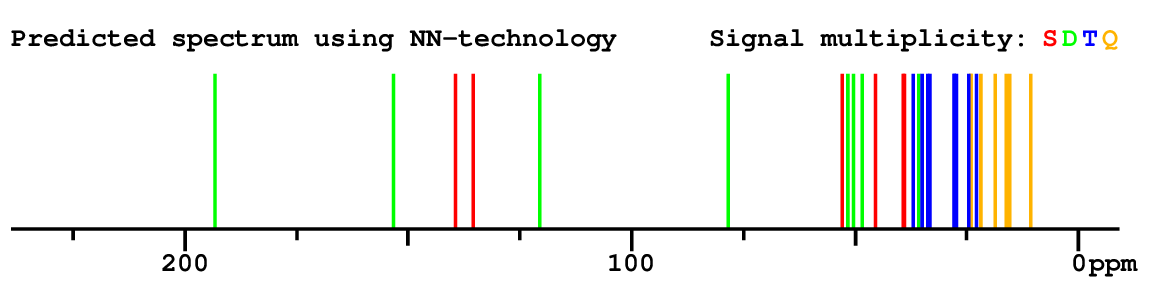
Structure Proposal # 2 CXPKYCXFLMPMEZ Deviation = 1.36ppm
Search WEB for skeleton Compound from PUBCHEM CSEARCH  |
|
|
|
|
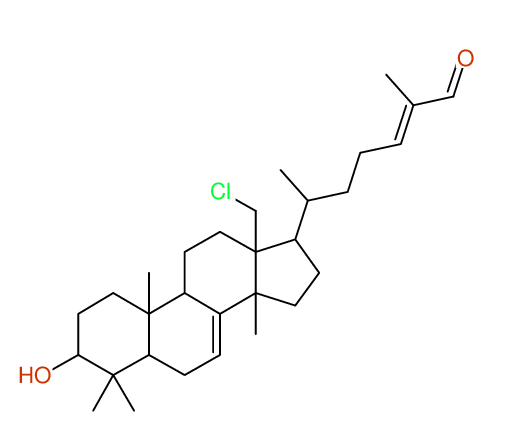
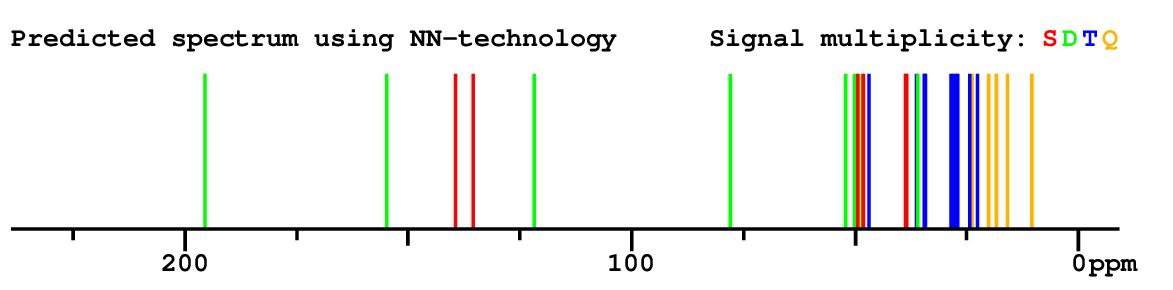
Structure Proposal # 3 BTZZXXSWVDVFPU Deviation = 1.41ppm
Search WEB for skeleton respect to your proposal |
|
|
|
|
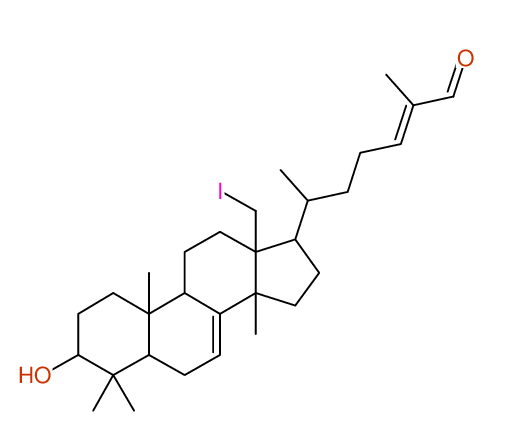
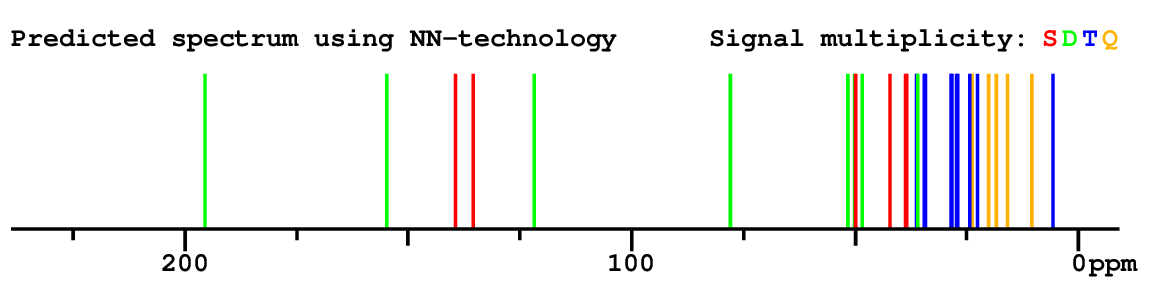
Structure Proposal # 4 BZQBLLHNQNUNPL Deviation = 1.42ppm
Search WEB for skeleton respect to your proposal |
|
|
|
|
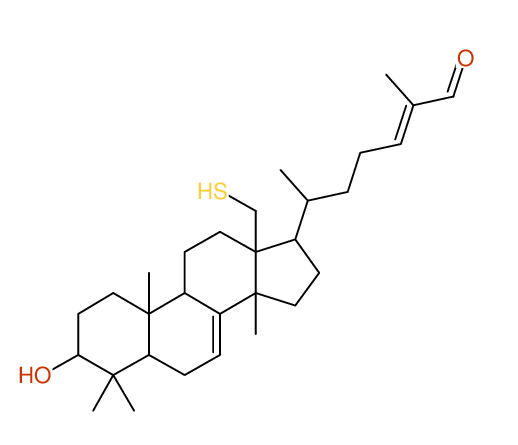
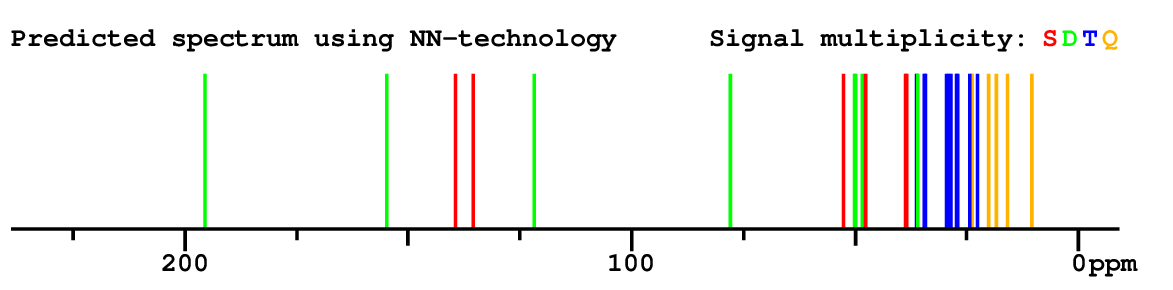
Structure Proposal # 5 XGZQCYMZSIVUIF Deviation = 1.43ppm
Search WEB for skeleton respect to your proposal |
|
|
|
|
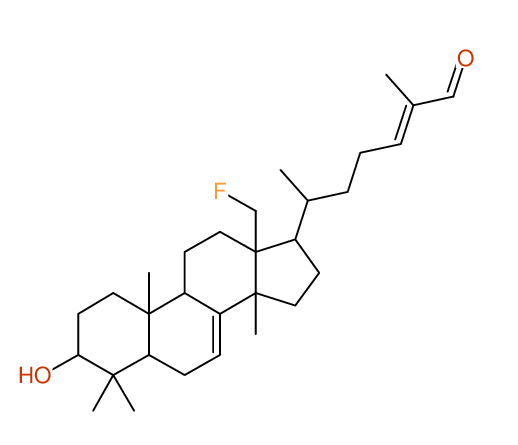
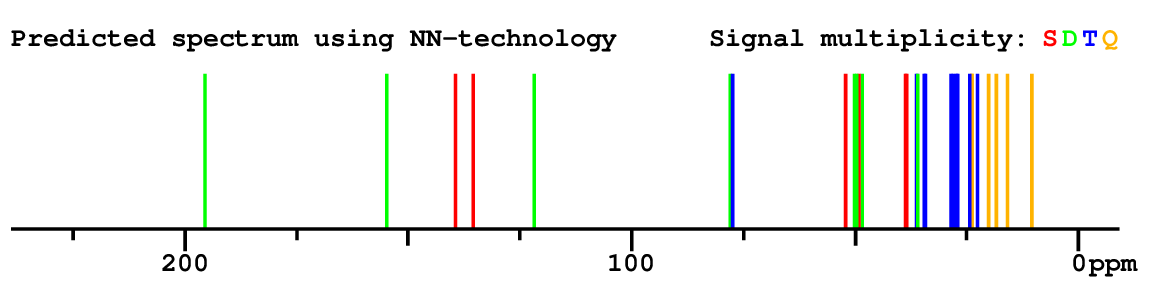
Structure Proposal # 6 JQWXHUKLYGXONB Deviation = 1.45ppm
Search WEB for skeleton respect to your proposal |
|
|
|
|
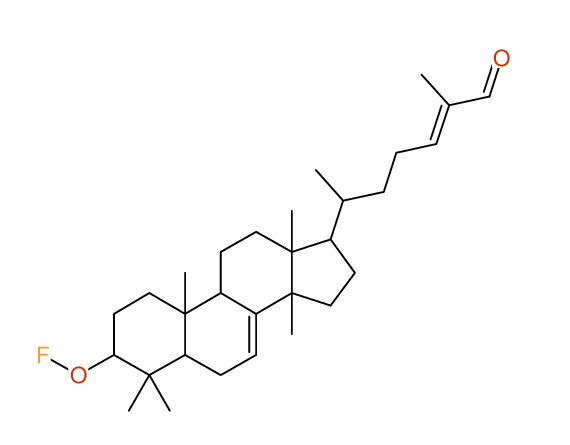
Structure Proposal # 7 ITVQGYBGLRDHKL Deviation = 1.49ppm
Search WEB for skeleton |
|
|
|
|
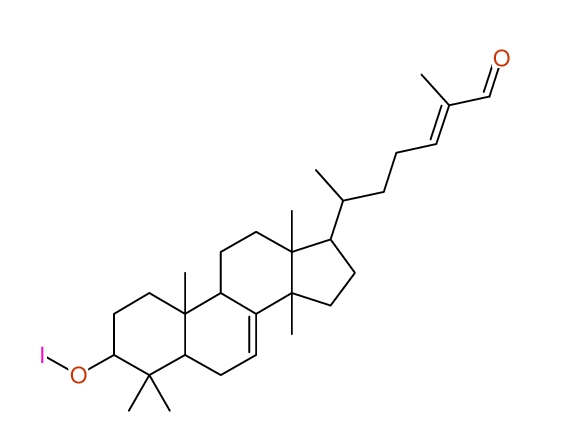
Structure Proposal # 8 OYOYPQYSRDKMDV Deviation = 1.51ppm
Search WEB for skeleton |
|
|
|
|
Structure Proposal # 9 LTUZTWKUZIDSQM Deviation = 1.54ppm
Search WEB for skeleton respect to your proposal |
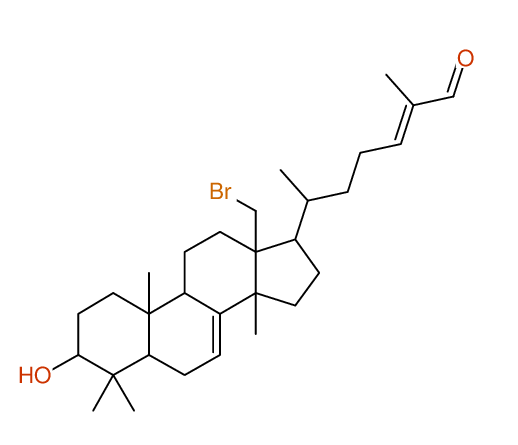
|
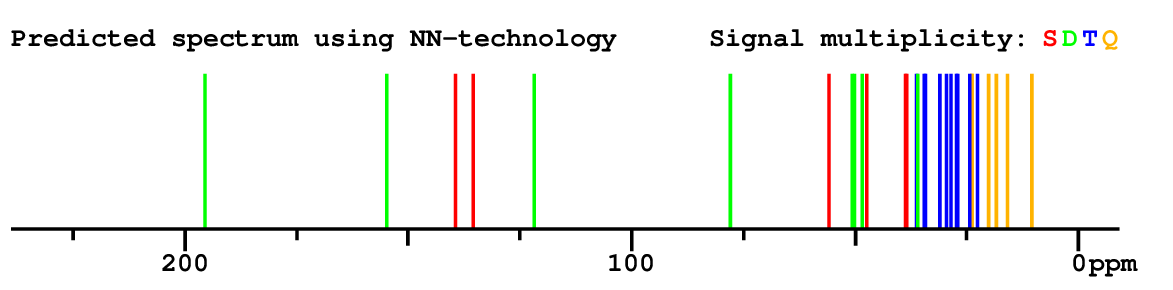 
|
|
|
Structure Proposal # 10 ISRFNVUZTSSBRB Deviation = 1.68ppm
Search WEB for skeleton |
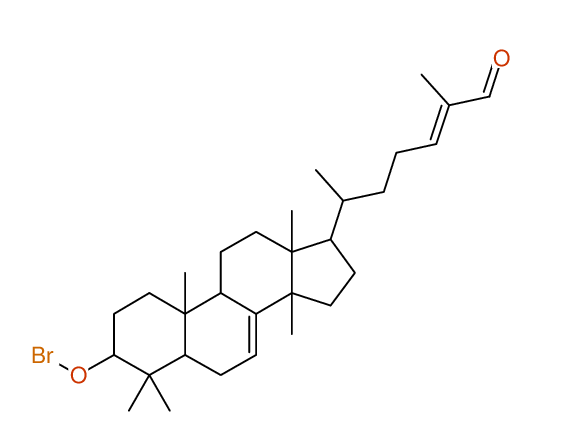
|
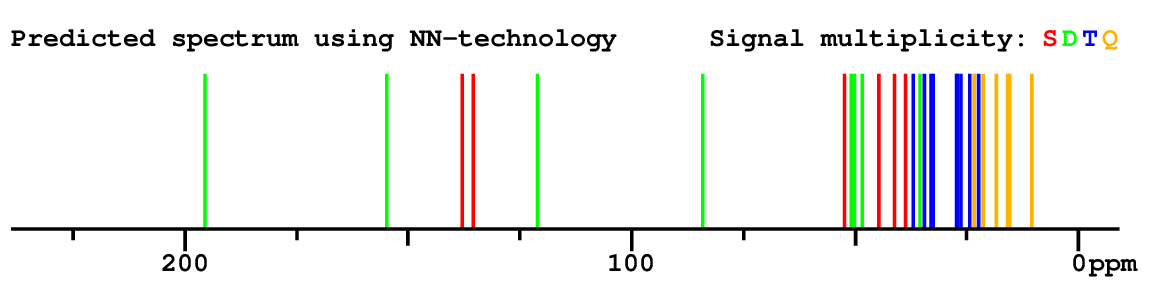 
|
|
|
Structure Proposal # 11 NBEYBYWRPQYABV Deviation = 1.80ppm
Search WEB for skeleton |
|
|
|
|
Structure Proposal # 12 PGUPFFLMLRDBMR Deviation = 1.99ppm
Search WEB for skeleton respect to your proposal |
|
|
|
|
Structure Proposal # 13 GQHAETXLIKAOGV Deviation = 2.20ppm
Search WEB for skeleton |
|
|
|
|
Structure Proposal # 14 UWCXETYNSOUPNN Deviation = 2.28ppm
Search WEB for skeleton |
|
|
|
|
Structure Proposal # 15 FNEQDTVVQMHZEW Deviation = 2.50ppm
Search WEB for skeleton |
|
|
|
|
Structure Proposal # 16 WCLIMMJBDLEKEH Deviation = 2.68ppm
Search WEB for skeleton |
|
|
|
|
Structure Proposal # 17 FFQRFMIXWDPKQJ Deviation = 2.78ppm
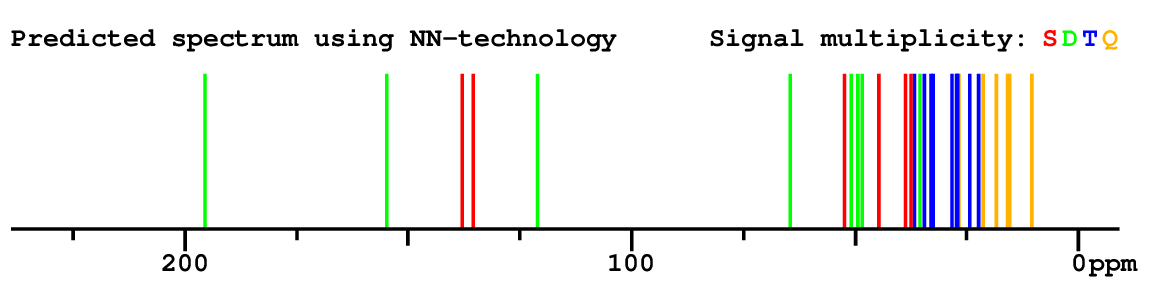
Search WEB for skeleton |
|
|
|
|
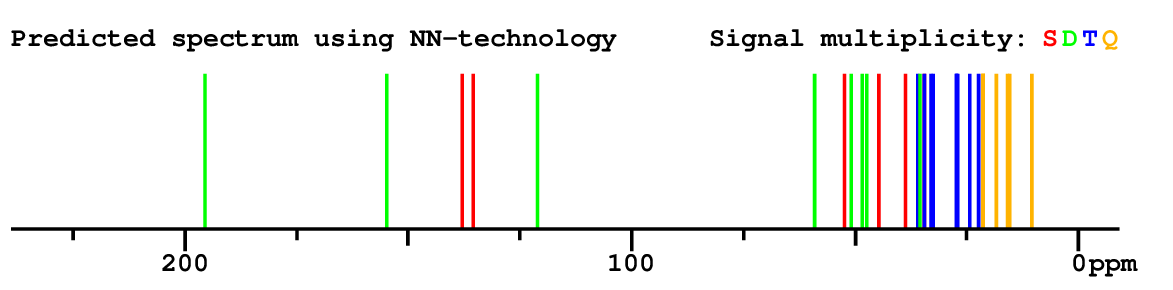
Structure Proposal # 18 OERAFRXLWPKWFS Deviation = 2.83ppm
Search WEB for skeleton |
|
|
|
|
Structure Proposal # 19 KGWDENSKEOZLCE Deviation = 3.02ppm
Search WEB for skeleton |
|
|
|
|
Structure Proposal # 20 DBLLDZISDXSOQE Deviation = 3.16ppm
Search WEB for skeleton |
|
|
|
|
Structure Proposal # 21 KAZPSMVKKRYKSL Deviation = 3.18ppm
Search WEB for skeleton respect to your proposal |
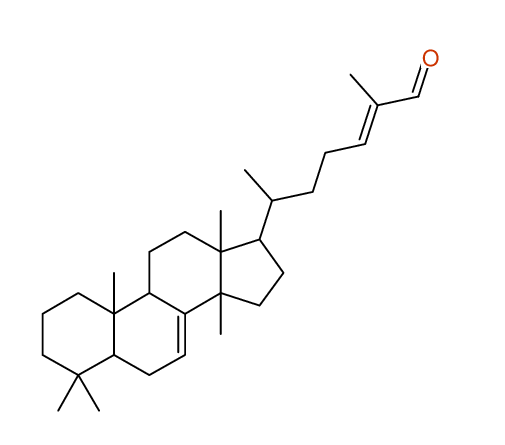
|
|
|
|
Structure Proposal # 22 FAGZXOGTBZVTDU Deviation = 3.67ppm
Search WEB for skeleton |
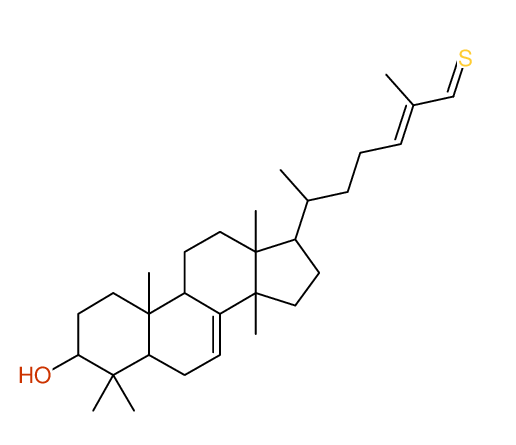
|
|
|
|
Structure Proposal # 23 BOUIWQLWVOGGOW Deviation = 3.82ppm
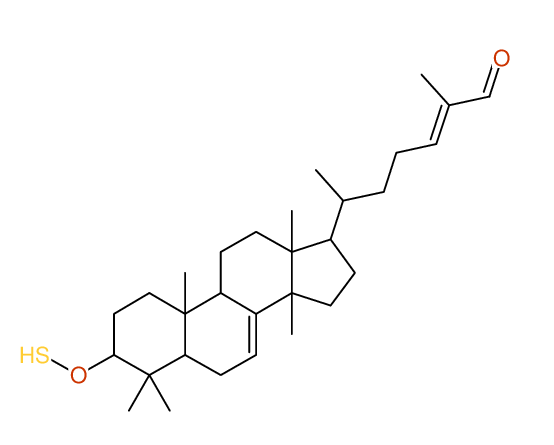
Search WEB for skeleton |
|
|
|
|
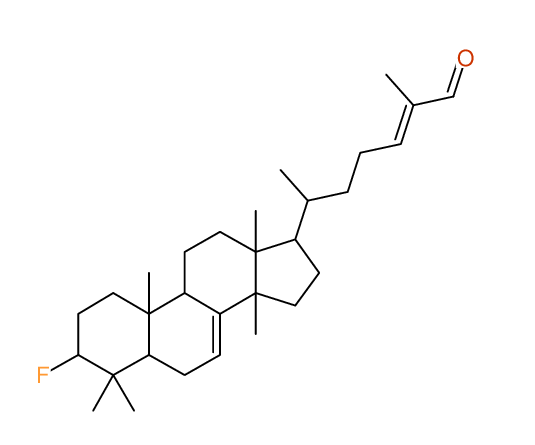
Structure Proposal # 24 LUPQORCXUOLGGQ Deviation = 3.82ppm
Search WEB for skeleton |
|
|
|
|
Structure Proposal # 25 NJCHPSBPVMEGHH Deviation = 3.83ppm
Search WEB for skeleton |
|
|
|
|
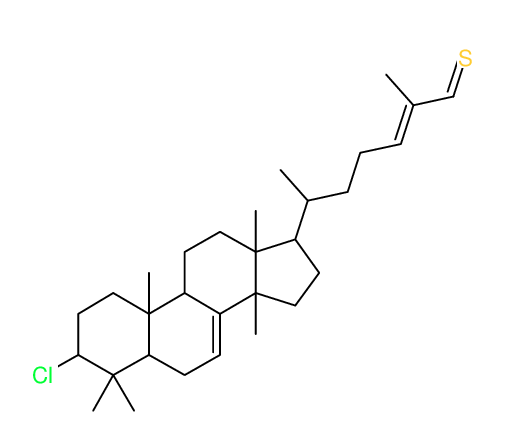
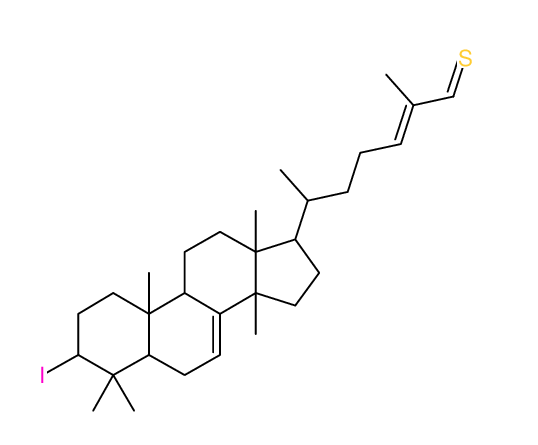
Structure Proposal # 26 KMRCGINCWKIDNV Deviation = 4.03ppm
Search WEB for skeleton |
|
|
|
|
Structure Proposal # 27 WJNMPAIRUXRZQK Deviation = 4.38ppm
Search WEB for skeleton |
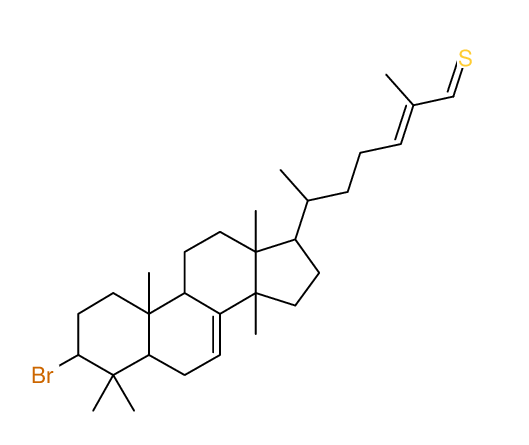
|
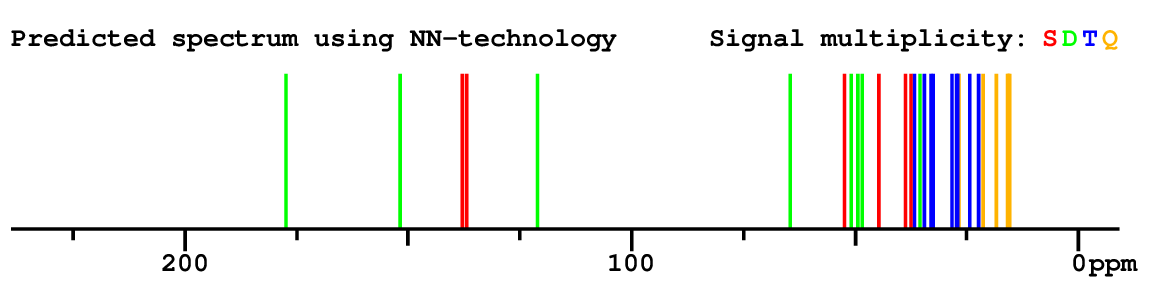 
|
|
|
Structure Proposal # 28 CDTBYFOLYXLSLM Deviation = 4.50ppm
Search WEB for skeleton |
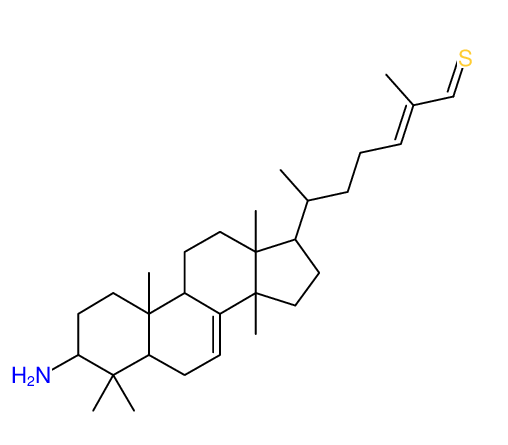
|
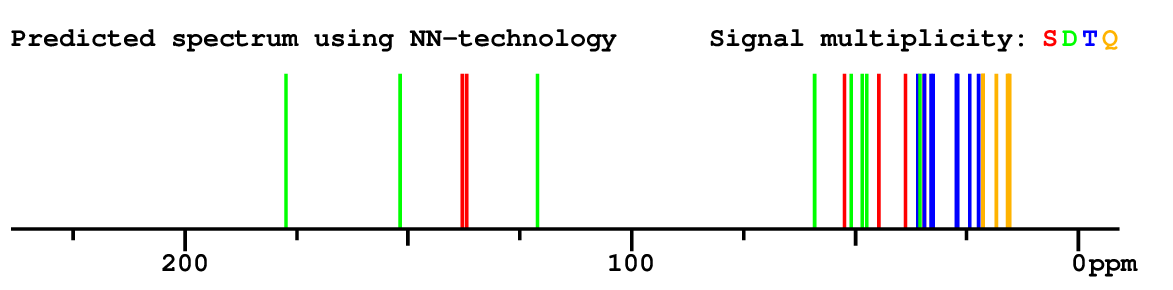 
|
|
|
Structure Proposal # 29 XGFRBFLVAUNUDH Deviation = 4.87ppm
Search WEB for skeleton |
|
|
|
|
Structure Proposal # 30 CXRMPSCVHCHGJV Deviation = 5.10ppm
Search WEB for skeleton |
|
|
|
|
Structure Proposal # 31 RWJULWKDECZOMH Deviation = 5.45ppm
Search WEB for skeleton |
|
|
|
|
Structure Proposal # 32 KDAKWQOIKLMZTC Deviation = 5.51ppm
Search WEB for skeleton respect to your proposal Compound from PUBCHEM CSEARCH  |
|
|
|
|
Structure Proposal # 33 BSWJBECZHYKCTG Deviation = 5.64ppm
Search WEB for skeleton |
|
|
|
|
Structure Proposal # 34 CHWNPXDVWCVSKK Deviation = 5.81ppm
Search WEB for skeleton respect to your proposal |
|
|
|
|
Structure Proposal # 35 DXOVMRHSLUIOJW Deviation = 5.85ppm
Search WEB for skeleton |
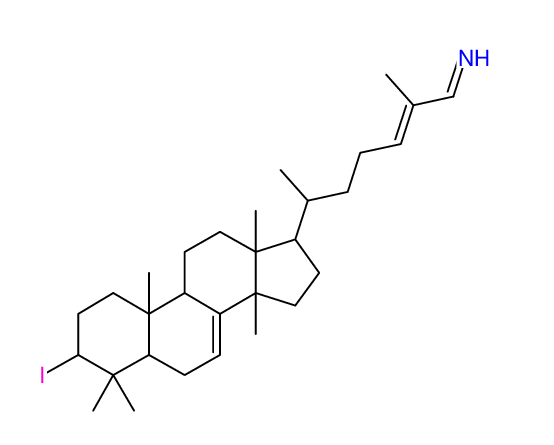
|
|
|
|
Structure Proposal # 36 QRBSZQIIDYJOON Deviation = 5.98ppm
Search WEB for skeleton |
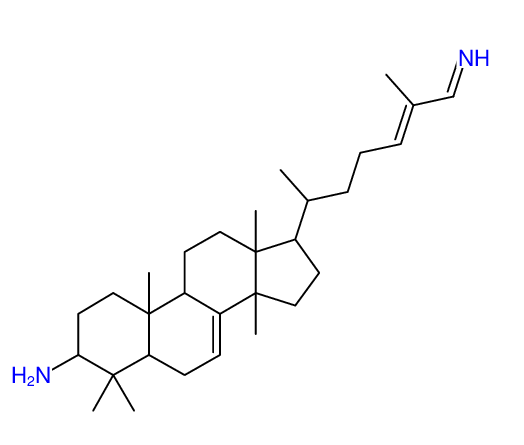
|
|
|
|
Structure Proposal # 37 IIDXGWPJQGQUCE Deviation = 6.08ppm
Search WEB for skeleton |
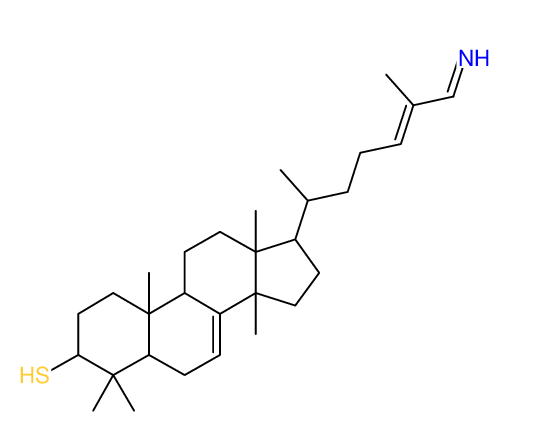
|
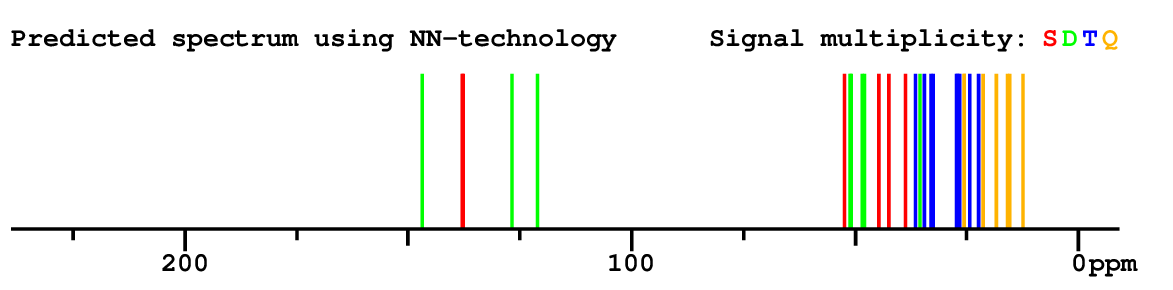 
|
|
|
Structure Proposal # 38 PDVSDPFTFYYSGD Deviation = 6.13ppm
Search WEB for skeleton |
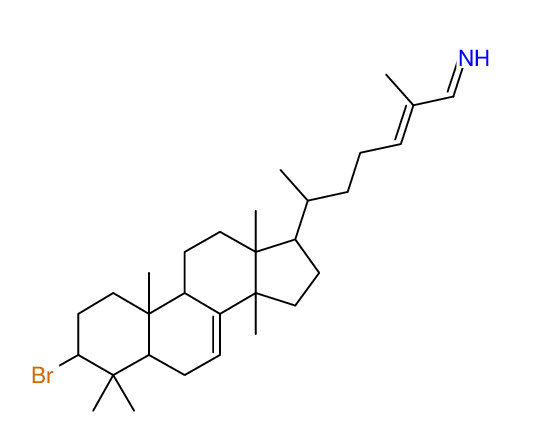
|
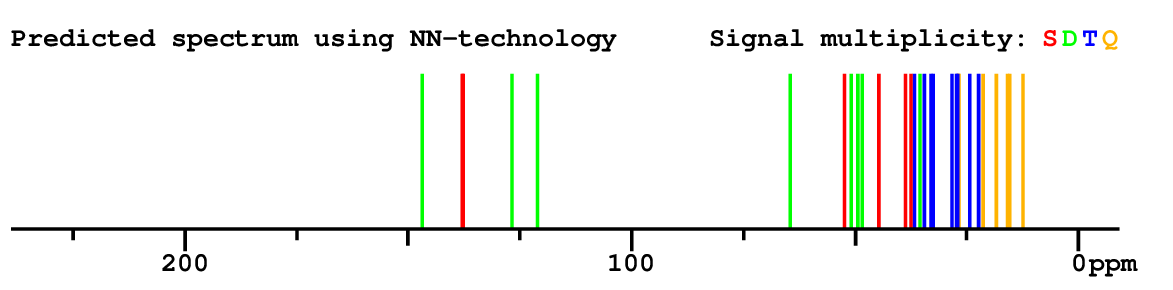 
|
|
|
Structure Proposal # 39 DICCPNLDOZNSML Deviation = 6.61ppm
Search WEB for skeleton respect to your proposal Compound from PUBCHEM CSEARCH  |
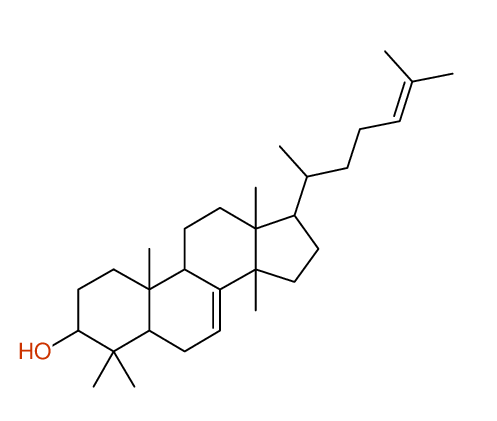
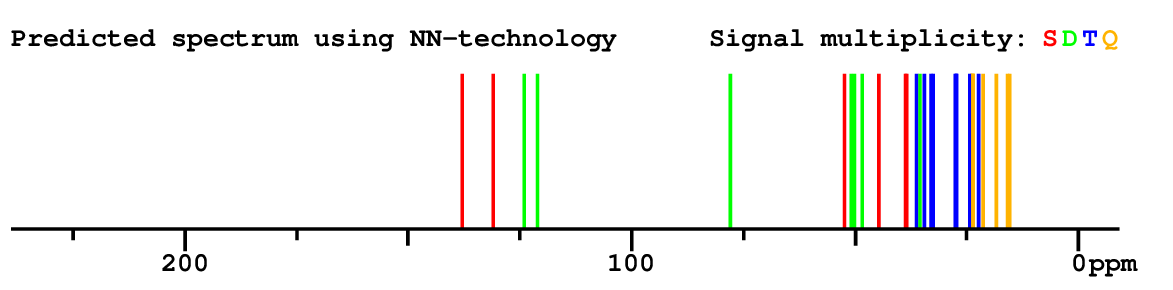
|
|
|
|
Structure Proposal # 40 ZBNIJWBROXKJHV Deviation = 6.79ppm
Search WEB for skeleton |
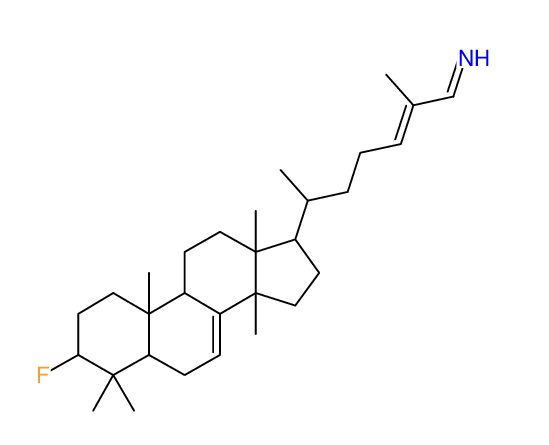
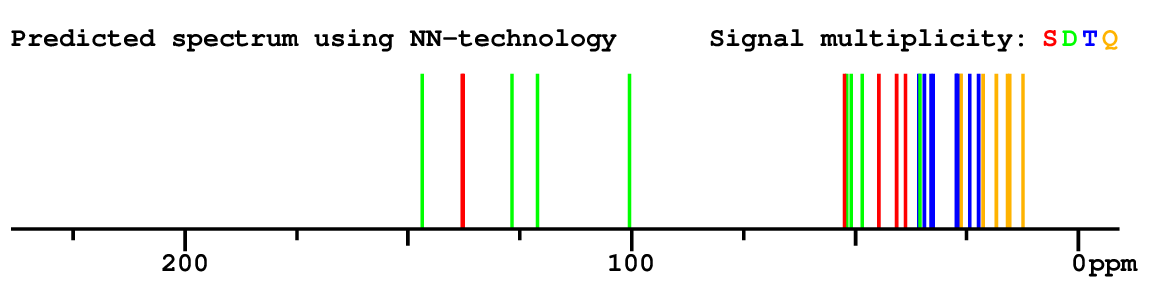
|
|
|
|
Structure Proposal # 41 OEJWNZLJHZJYTI Deviation = 7.18ppm
Search WEB for skeleton respect to your proposal |
|
|
|
|
Proposal # 1 Similarity: 1.36ppm CXPKYCXFLMPMEZ PUBCHEM 
|
Proposal # 2 Similarity: 1.49ppm ITVQGYBGLRDHKL 
|
Proposal # 3 Similarity: 1.51ppm OYOYPQYSRDKMDV 
|
|
Proposal # 4 Similarity: 1.68ppm ISRFNVUZTSSBRB 
|
Proposal # 5 Similarity: 1.80ppm NBEYBYWRPQYABV 
|
Proposal # 6 Similarity: 2.20ppm GQHAETXLIKAOGV 
|
|
Proposal # 7 Similarity: 2.28ppm UWCXETYNSOUPNN 
|
Proposal # 8 Similarity: 2.50ppm FNEQDTVVQMHZEW 
|
Proposal # 9 Similarity: 2.68ppm WCLIMMJBDLEKEH 
|
|
Proposal # 10 Similarity: 2.78ppm FFQRFMIXWDPKQJ 
|
Proposal # 11 Similarity: 2.83ppm OERAFRXLWPKWFS 
|
Proposal # 12 Similarity: 3.02ppm KGWDENSKEOZLCE 
|
|
Proposal # 13 Similarity: 3.16ppm DBLLDZISDXSOQE 
|
Proposal # 14 Similarity: 3.67ppm FAGZXOGTBZVTDU 
|
Proposal # 15 Similarity: 3.82ppm BOUIWQLWVOGGOW 
|
|
Proposal # 16 Similarity: 3.82ppm LUPQORCXUOLGGQ 
|
Proposal # 17 Similarity: 3.83ppm NJCHPSBPVMEGHH 
|
Proposal # 18 Similarity: 4.03ppm KMRCGINCWKIDNV 
|
|
Proposal # 19 Similarity: 4.38ppm WJNMPAIRUXRZQK 
|
Proposal # 20 Similarity: 4.50ppm CDTBYFOLYXLSLM 
|
Proposal # 21 Similarity: 4.87ppm XGFRBFLVAUNUDH 
|
|
Proposal # 22 Similarity: 5.10ppm CXRMPSCVHCHGJV 
|
Proposal # 23 Similarity: 5.45ppm RWJULWKDECZOMH 
|
Proposal # 24 Similarity: 5.64ppm BSWJBECZHYKCTG 
|
|
Proposal # 25 Similarity: 5.85ppm DXOVMRHSLUIOJW 
|
Proposal # 26 Similarity: 5.98ppm QRBSZQIIDYJOON 
|
Proposal # 27 Similarity: 6.08ppm IIDXGWPJQGQUCE 
|
|
Proposal # 28 Similarity: 6.13ppm PDVSDPFTFYYSGD 
|
Proposal # 29 Similarity: 6.79ppm ZBNIJWBROXKJHV 
|
|
Proposal # 1 Similarity: 1.36ppm CXPKYCXFLMPMEZ PUBCHEM 
|
Summary of searching for alternative Structures
Project: CHEM.PHARM.BULL.,62,937[2014]CSEARCH-NUMBER:UWBT013934-20160521 | ||||||||||||||||
| ||||||||||||||||
| Your proposal | Best proposal | Best proposal (out of 1) which exists either in CSEARCH or PUBCHEM |
||||||||||||||
|
Deviation = 1.36 ppm Position = 1 |
Deviation = 1.35 ppm |
Deviation = 1.36 ppm Position = 1 |
||||||||||||||
 |
 |
 |
||||||||||||||
| ||||||||||||||||